NEMAR and NSG
Processing NEMAR data using The Neuroscience Gateway
NEMAR is a new category of data science facility, a public integrated data, tools, and compute resource (datcor). Multiple software environments are widely used for analysis of human neuroelectromagnetic (NEM) data. Rather than building a seamless compute facility based on only one such environment, NEMAR is a collaboration with the developers of The Neuroscience Gateway (NSG, www.nsgportal.org) at the San Diego Supercomputer Center, a neuroscience community resource that supports multiple software environments (MATLAB, python, R), and toolsets (EEGLAB, Open Brain, Freesurfer, etc.) – enabling users to submit analysis scripts of their own creation operating in any NSG-supported environment to process NEMAR/OpenNeuro NEM datasets on NSF-supported high-performance computing resources. NSG supports progress in the field of electromagnetic brain imaging by freely supporting the use of computational processing methods that require or can benefit from the availability of substantial computational power.
Bypassing the data transfer bottleneck. NEMAR encourages intensive exploration of NEM datasets that have been made public by their authors by uploading them to OpenNeuro (OpenNeuro.org). Giving NSG analysis scripts direct access to data housed in NEMAR allows users to bypass the slow, fragile, and computationally and energywise costly processes of downloading NEMAR data of interest and then again uploading the data to NSG or elsewhere for analysis. NEM datasets in OpenNeuro are copied to NEMAR where they are further curated, their quality is assessed, and they are then made available within NEMAR, for user search, inspection, and open-ended analysis. To help users assess the potential value to their research of the available datasets, NEMAR provides some data quality measures and visualization options.
The NEMAR use process is simple: (1) Use the NEMAR data search and visualisation tools to identify one or more BIDS-formatted NEM datasets of interest. (2) Then write an NSG data processing script citing the control number(s) of the dataset(s) to be processed (example: ds000123) - as demonstrated below. The NSG processing script may use any software environment and tools that NSG supports. (3) When processing is complete, the user will be notified by email that the results specified in the data processing script are (4) available for download.
Freely processing NEMAR data using high-performance computing. The Neuroscience Gateway (NSG) allows neuroscientists to process and model data, run simulations, and train AI/ML networks on a nationally supported high-performance supercomputer network on which a variety of neuroscience data processing and modelling software are available. The NSG compute resource is openly and freely available to researchers working on not-for-profit projects. NSG provides an easy-to-use web portal-based user environment as well as programmatic access via a REST software interface. Users of the EEGLAB software environment, in particular, may use a set of REST-based EEGLAB tools (nsgportal) to launch, monitor, and examine results of NSG jobs directly from the EEGLAB menu window.
Before beginning to use NEMAR for data analysis, request an NSG account on the NSG registration page.
Processing NEMAR data via NSG – Overview
Using an EEGLAB MATLAB script Using a Python script
Using the desktop EEGLAB nsgportal interface Frequently Asked Questions (FAQ)
Using a MATLAB script (non-EEGLAB) References
Questions/Suggestions: Email the NEMAR team at support@nemar.org
Processing NEMAR datasets using Matlab or Python NSG data analysis scripts
Note 1: Avoid timeouts. There is a 48-hour NSG job timeout limit -- your NSG script must require less time to run than this. If your task is more extensive than this, have your analysis script produce and return intermediate results.
Note 2: Avoid large data downloads. By default, on completion of the processing you specify in your analysis script NSG will download the script’s complete working directory/folder; by default this will contain the whole input data that it processes, along with any files your script creates. If you do NOT want to receive a large download including the whole dataset(s) you have just processed, you must explicitly remove the input data and any other files you do not want to receive within your NSG processing script. You can always perform further processing on the same data again using NSG, and if you wish your further analysis can include any earlier intermediate results you may upload along with the new processing script.
HOW TO PROCESS NEMAR DATA ON MATLAB
To use a MATLAB script to process NEMAR data via NSG:
- Copy one of the linked sample MATLAB scripts to a text editor.
- Set the name of the NEMAR dataset you wish to process in place of ‘bidsNAME’ below - NEMAR dataset names are of the form ‘ds00xxxx’.
- Create a new folder to hold the job data. Give this folder any name you wish.
- Save the completed script as ‘input.m’ within the new folder.
- Create a zip archive of the new folder.
- Note: if you instead zip the script itself, NSG will return an error.
- Select “Data” in the left menu.
- Press the “Upload” button to upload the zip file above.
- Select “Task” in the left menu.
- Press the “Create New Task” button.
- Enter a description (optional).
- Press the “Select Input Data” and select the zip file you have just uploaded.
- Select “Tools” and select ‘EEGLAB’ or ‘MATLAB’.
- Optionally, select “Select Parameters” (typically the default parameters are sufficient).
- Select “Save and Run Task”.
- Your job will be queued for processing; NSG will inform you by email when its processing is complete. See the NSG Tutorials for more information.
The following sample NEMAR scripts demonstrate the use of different processing environments:
NSG hosts the extensive EEGLAB tool environment running on MATLAB. To write an EEGLAB script to process a NEMAR dataset, modify the code example below and run your script as follows:
The sample NSG script for EEGLAB below demonstrates how to process one or more NEMAR datasets via the Neuroscience Gateway (NSG) using the EEGLAB environment:
%
% SAMPLE NSG MATLAB SCRIPT USING EEGLAB TO PROCESS A NEMAR/OPENNEURO DATASET
%
bidsName = 'ds??????'; % The NEMAR/OpenNeuro ID of the dataset you want to process
%
% Start up EEGLAB within the NSG session
%
eeglab; % Open the EEGLAB environment
%
% Build the NEMAR address of the selected dataset
%
filePath = [ getenv('NEMARPATH') bidsName ]; % NEMARPATH is a global variable in NSG MATLAB
% pointing to the NEMAR data directory.
[status cwd] = system('pwd'); % Get the current working directory on NSG.
cwd = strip(cwd); % Remove whitespace from path directory under the pwd
out_dir = fullfile(cwd, 'eegbidsout'); % in which to put the EEGLAB Study files.
%
[STUDY,EEG] = pop_importbids(filePath, 'outputdir', out_dir); % Import the BIDS dataset
% as an EEGLAB STUDY
% in the named subdir.
%
% Test example: Here simply query the imported STUDY and print some results into an ascii file.
%
fid = fopen('results.txt', 'w'); % Open a text file to store dataset information returned below
%
fprintf(fid,'***************************\n'); % Print to an output information file:
fprintf(fid,'BIDS dataset %s\n', bidsName); % - the OpenNeuro/NEMAR dataset ID
fprintf(fid,'***************************\n');
fprintf(fid,'%d datasets\n', length(EEG)); % - the total number of EEG data files
fprintf(fid,'%d to %d channels\n', ...
min([EEG.nbchan]), max([EEG.nbchan])); % - their range of EEG channel numbers
fprintf(fid,'%d to %d seconds\n', ...
round(min([EEG.pnts])/EEG(1).srate), round(max([EEG.pnts])/EEG(1).srate));
% - their range of durations in sec
fprintf(fid,'%d to %d events\n', ...
min(cellfun(@length, {EEG.event})), max(cellfun(@length, {EEG.event})));
% - their range of event numbers
fclose(fid); % Close the ascii information file
type('results.txt'); % Print the name of the output information file as a reminder
%
% NOTE: By default, the EEGLAB study
% converted from the BIDS repository will be saved for download
% ALONG WITH any results files your script has created.
% When you do NOT need to download the STUDY data files (which may be large),
% but only the results derived by this script,
% MAKE SURE TO DELETE all the STUDY files (as below) before ending your script.
%
rmdir(out_dir, 's') % Remove the imported STUDY files to avoid a large unneeded download
To run this script: As detailed above, login to NSG, identify yourself as an EEGLAB user and upload the script. It will run in batch mode; NSG will inform you by email when processing is complete.
- Processing NEMAR data directly from an EEGLAB session using the EEGLAB NSGportal plug-in interface
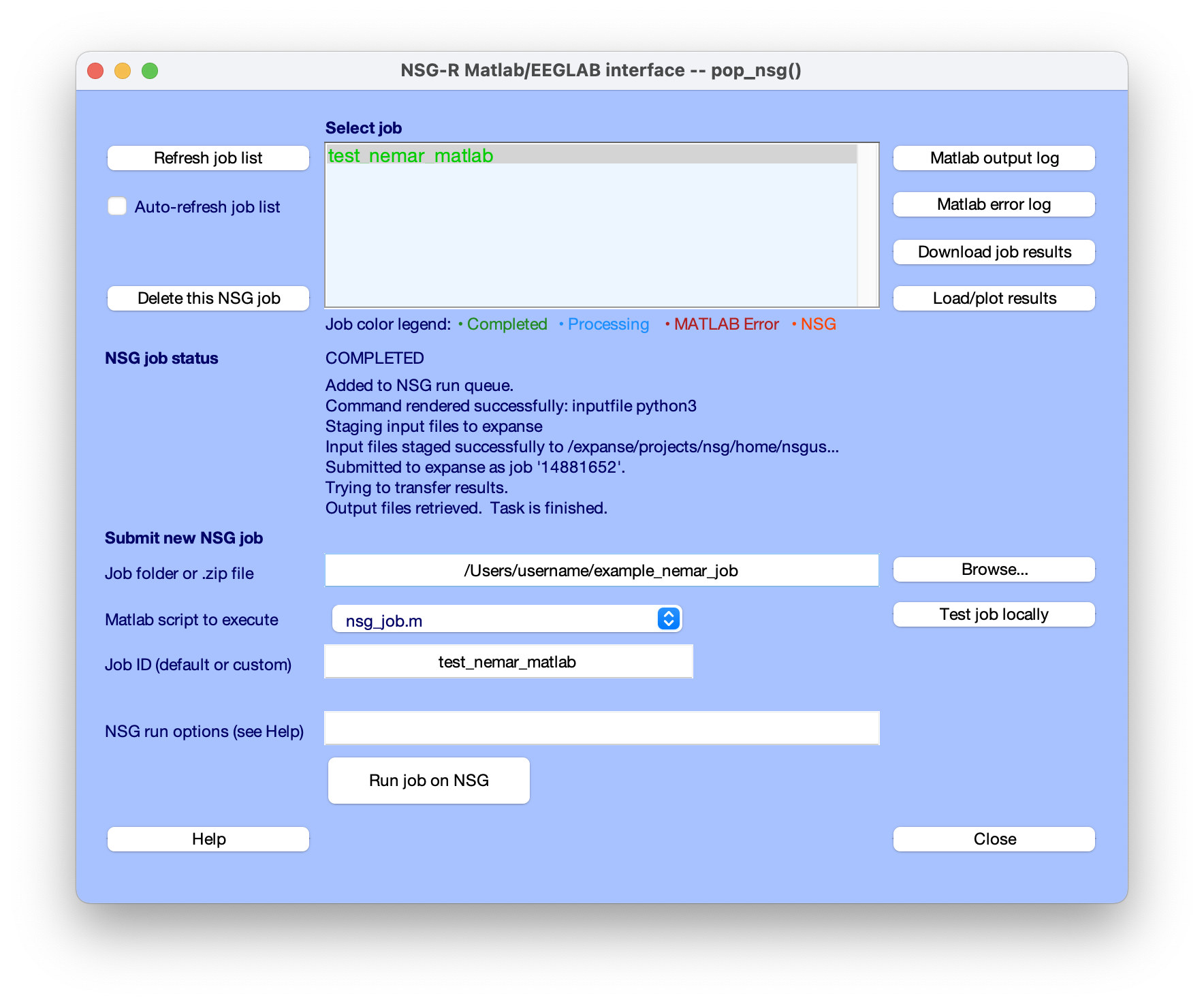
EEGLAB users can also take advantage of the EEGLAB nsgportal plug-in to submit a processing script to NSG directly (using the NSG REST language interface), to monitor its progress, and to retrieve and visualize (as/when relevant) its results - all directly through the EEGLAB menu. A paper by Martinez-Cancino et al. provides a clear tutorial on using the EEGLAB nsgportal plug-in. Below we show a screenshot from submitting the EEGLAB script as shown in Section 1 above to NSG and monitoring the results the via the EEGLAB nsgportal plug-in user interface.
Figure. The pop-up menu of the EEGLAB nsgportal plug-in (access using menu link Tools/NSG Portal). The middle of the window logs the progress of the submitted job running the NEMAR processing script (1) above (‘test nemar matlab’). See Martinez-Cancino et al. for more detail.
- Processing NEMAR data directly using MATLAB (outside of EEGLAB)
Processing NEMAR datasets using MATLAB outside of the EEGLAB software environment is possible as well. Your MATLAB script will need to copy the BIDS dataset to the local working directory and then operate directly on the BIDS file hierarchy.
%
% SAMPLE NSG SCRIPT RUNNING ON MATLAB TO OPERATE DIRECTLY ON A NEMAR DATASET (OUTSIDE OF EEGLAB)
%
bidsName = 'ds002691'; % The NEMAR/OpenNeuro control ID of the dataset you want to process.
%
% Build the NEMAR address of the selected dataset
%
filePath = [ getenv('NEMARPATH') bidsName ]; % NEMARPATH is a global variable in NSG MATLAB
% pointing to the dir in which the NEMAR data are stored.
[status cwd] = system('pwd'); % Get the current working directory on NSG.
cwd = strip(cwd); % Remove any whitespace from the pathname.
out_dir = fullfile(cwd, 'eegbids'); % The dir under pwd in which to put the dataset BIDS files.
status = copyfile(filePath, out_dir); % Copy the BIDS dataset folder hierarchy to the local working directory
%
% Now operate directly on the BIDS DATASET files as you wish …
%
% [your code goes here]
%
% Again, remember to EXPLICITLY REMOVE THE LOCAL DATASET COPY
% if/when you do not want the whole dataset to be downloaded
% with the results files your script may create.
%
system(['chmod -R u+w ' out_dir]); % Make sure permission is correct for successful deletion.
rmdir(out_dir, 's') % Remove the copied dataset files to avoid a large download.
- Processing NEMAR data using Python
Processing NEMAR datasets using Python is also possible. You can copy the BIDS dataset to the local working directory and then operate directly on the BIDS file as in the MATLAB example above.
|
import os
shutil.copytree(DATASETDIR, copiedDir) # Copy dataset to current working directory before processing |
Frequently Asked Questions (FAQ)
Can I also process my own data on NSG?
Yes; you will need to include the data in the zip file you upload to NSG. BUT – if your data are in BIDS format and are publicly available on OpenNeuro/NEMAR, then you can process the data using scripts based on the above examples without needing to re-upload the data.
Can I train deep learning models on EEG datasets on NSG?
Yes. NSG also has a GPU time allocation, and serves both TensorFlow and PyTorch.
Can I process multiple BIDS datasets at once on NSG?
Yes, simply edit your script accordingly to import and process the datasets. NEMAR doesn’t actually copy the datasets to the working directory – though your script may do so if it converts them to (e.g.) EEGLAB STUDYs. Note 7/14/22: We are working to allow NEMAR search to identify individual participants and runs (aka ‘scans’) of interest, as well as specific event-related data epochs (across datasets) for analysis. When these features are ready (or even now, using your own custom conversion and processing code), working across multiple NEMAR datasets may become more space-efficient.
Is there a limit to the amount of data I can process on NSG?
There is currently no fixed limit in terms of the (reasonable) amount of data processing you can perform. You will be contacted if the NSG staff notice you are using an extraordinary amount of resources.
Is there a limit to the amount of RAM and/or processing time on NSG?
Standard NSG limits apply. Jobs can typically run only for a maximum of 48 hours on the Expanse supercomputer at SDSC. On Expanse, the maximum amount of RAM your script can make use of is 243 GB.
Can I parallelize my job on NSG?
Yes, and this is highly recommended. You may use MATLAB parfor loops or their Python equivalent to run concurrently on up to 128 cores. However, we recommend using 32 or 64 cores only, as each core will need some RAM to process data and your process may quickly run out of RAM (even given 243 GB), at which point it will be killed by the NSG scheduler.
My zipfile of results returned by NSG is huge. Why?
By default, the dataset your script copied, or EEGLAB STUDY your script converted from the NEMAR BIDS dataset will be saved in the working directory and included in the download along with any result files it may create. If you do not need to download the STUDY your script has processed (which may be quite large!!) make sure to delete all the dataset BIDS or STUDY files before ending your script! See examples in the sample scripts above.
Can I use NSG if I am working for a commercial company?
The short answer is no. As per standard NSG policy, you will not be able to create an NSG account unless you are working on a not-for-profit project.
Can I use NSG if I am not affiliated with a university or non-profit?
Again, standard NSG policies apply. The short answer is probably yes - if your project is nonprofit and open research oriented.
Can I use NSG if I am not working in the US or for a US institution?
Again, standard NSG policies apply. The short answer is yes. NSG is open to the world (with a very few exceptions).
Is there a limit to the amount of data I can process on the supercomputer?
Again, standard NSG policies apply. There is no hard limit. NSG admin will contact you personally to check on your project if you begin to use an inordinate amount of resources compared to other users.
Is there a limit to the processing time I can use on the supercomputer?
Here again, standard NSG policies apply, and there is no hard limit. NSG admin will contact you personally to check on your project if you begin to use an inordinate amount of resources compared to other users. However, as indicated above there is a hard limit to the length of time an NSG script can run – at 48 hours it will be killed by the scheduler.
The NEMAR/NSG facility is really helpful to me. Is there anything I can do for NSG and NEMAR?
Yes, please credit NEMAR and cite our papers in your work. Also, we plan to begin a NEMAR newsletter and will be soliciting user stories - offer to work with us on one!
References
NEMAR. Delorme, A., Truong, D., Youn, C., Sivagnanam, S., Yoshimoto, K., Poldrack, R.A., Majumdar, A. and Makeig, S., 2022. NEMAR: An open access data, tools, and compute resource operating on NeuroElectroMagnetic data. arXiv preprint arXiv:2203.02568.
NSG. S Sivagnanam, A Majumdar, K Yoshimoto, V Astakhov, A Bandrowski, M. E. Martone, and N. T. Carnevale. Introducing the Neuroscience Gateway, IWSG, volume 993 of CEUR Workshop Proceedings, CEUR-WS.org, 2013.
BIDS. Gorgolewski, K.J., Auer, T., Calhoun, V.D., Craddock, R.C., Das, S., Duff, E.P., Flandin, G., Ghosh, S.S., Glatard, T., Halchenko, Y.O. and Handwerker, D.A., 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific data, 3(1), pp.1-9.
BIDS-EEG. Pernet, C.R., Appelhoff, S., Flandin, G., Phillips, C., Delorme, A. and Oostenveld, R., 2019. BIDS-EEG: an extension to the Brain Imaging Data Structure (BIDS) Specification for electroencephalography.
BIDS-MEG. Niso, G., Gorgolewski, K.J., Bock, E., Brooks, T.L., Flandin, G., Gramfort, A., Henson, R.N., Jas, M., Litvak, V., T Moreau, J. and Oostenveld, R., 2018. MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Scientific data, 5(1), pp.1-5.
BIDS-iEEG. Holdgraf, C., Appelhoff, S., Bickel, S., Bouchard, K., D’Ambrosio, S., David, O., Devinsky, O., Dichter, B., Flinker, A., Foster, B.L. and Gorgolewski, K.J., 2019. iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Scientific data, 6(1), pp.1-6.
EEGLAB. Delorme, A. and Makeig, S., 2004. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. Journal of neuroscience methods, 134(1), pp.9-21.
EEGLAB-BIDS. Delorme, A., Truong, D., Martinez-Cancino, R., Pernet, C., Sivagnanam, S., Yoshimoto, K., Poldrack, R., Majumdar, A. and Makeig, S., 2021, May. Tools for Importing and Evaluating BIDS-EEG Formatted Data. In 2021 10th International IEEE/EMBS Conference on Neural Engineering (NER) (pp. 210-213). IEEE.
EEGLAB-NSG. Martínez-Cancino, R., Delorme, A., Truong, D., Artoni, F., Kreutz-Delgado, K., Sivagnanam, S., Yoshimoto, K., Majumdar, A. and Makeig, S., 2021. The open EEGLAB portal interface: High-performance computing with EEGLAB. NeuroImage, 224, p.116778.